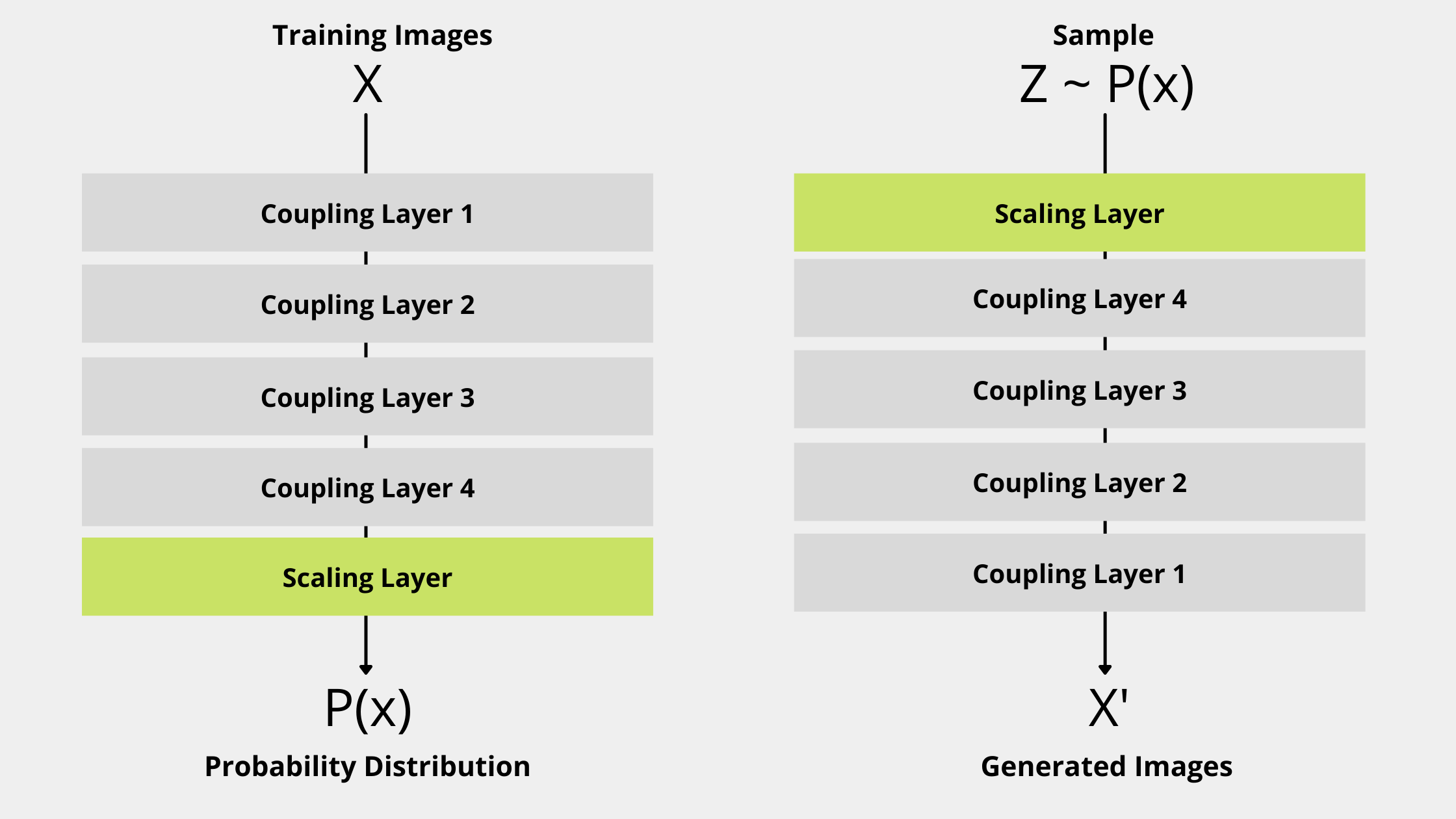
Motion detection in a video footage has always been an interesting topic among the Machine Learning enthusiasts. It has many purposes. Motion detection can be used for automatic video analysis in video surveillance systems. It can be used to capture movements on a wildlife camera. Another application is to detect number of vehicles passing by on a highway.
In this article, we will build a fully working motion detector. It will not only detect motion, but will also be able to track motion of different objects. By this process, we will learn a lot about image processing techniques in OpenCV. By the end, you will have all the codes required to build your own motion detector.
Problem Statement
We will build a Computer Vision model for motion detection. It will be able to detect moving people in a video footage. Each person detected will have a different colored bounding box. The motion’s path will be shown with the same color as that of the bounding box.
We will achieve motion detection by comparing each video frame to the previous one. The spots which are different from the last frame are detected. For the predicting motion’s path, Kalman Filter is used. The final result will look like this:
Coding Our Motion Detector with Python
Firstly, we look at and understand the code for the motion detection. Then, we write scripts for tracking the path of moving objects. We will use the scripts and draw path of the objects on the frame. So, let’s begin-
1. Reading the video from location
We’ve used cv2.VideoCapture to capture the video from a file location. You can change it to your own location.
import cv2
import imutils
import numpy as np
cap = cv2.VideoCapture('video.mp4')2. Preparing the frames
while cap.isOpened():
# Capture frame-by-frame
ret, frame = cap.read()
if ret is None:
break
try:
frame = imutils.resize(frame, width=750)
except:
break
gray = cv2.cvtColor(frame, cv2.COLOR_BGR2GRAY)
gray = cv2.GaussianBlur(gray, (21, 21), 0)In this block of code, we capture each frame of the video and resize it. Then, we convert it to gray scale image. Now the frame’s pixels have values in the range of 0-255. Later, the frame is blurred to smooth it out a little.
3. Detecting motion in the frames
We begin with setting the first frame if it is None. If the number of frames exceed 5, we reset the delay counter and set the first frame with next frame.Motion is detected by comparing first frame with the next frame. The difference between frames is calculated using absdiff method.
if first_frame is None:
first_frame = gray
next_frame = gray
delay_counter += 1
if delay_counter > 5:
delay_counter = 0
first_frame = next_frame
frame_delta = cv2.absdiff(first_frame, next_frame)
thresh = cv2.threshold(frame_delta, 25, 255, cv2.THRESH_BINARY)[1]
thresh = cv2.dilate(thresh, None, iterations=2)
cnts, _ = cv2.findContours(thresh.copy(), cv2.RETR_EXTERNAL, cv2.CHAIN_APPROX_SIMPLE)threshold() method is used to make the pixels white whose values are greater than threshold value 25. The frames are dilated and we finally find the contours where motion is detected.
This is what it looks like-
5. Finding centroids of the objects in motion
For each contour with contour area greater than 1000, we find its centroid and store it. Simultaneously, we also store the coordinates of bounding box the centroid belongs to.
centers = []
coords = []
for c in cnts:
try:
(x, y, w, h) = cv2.boundingRect(c)
if cv2.contourArea(c) > 1000:
b = np.array([[x + w // 2], [y + h // 2]])
centers.append(np.round(b))
coords.append([x, y, x + w, y + h])
except ZeroDivisionError:
passWe have successfully detected bounding boxes and their centroids for the motion detected in the video footage. So, the first half of our task is completed. Now, we proceed towards tracking the objects in motion.
Kalman Filter
Kalman filter algorithm estimates the states of a system given the observations or measurements. It is a useful tool for a variety of different applications including object tracking and autonomous navigation systems, economics prediction, etc.
The Kalman filter is essentially a set of mathematical equations that implement a predictor-corrector type estimator that is optimal in the sense that it minimizes the estimated error covariance when some presumed conditions are met [1].
Mathematical Equation [1]:
The Kalman filter addresses the general problem of trying to estimate the state x ∈ ℜn of a discrete-time controlled process that is governed by the linear stochastic difference equation
xk = Axk-1 + Buk + wk-1
with a measurement m y ∈ℜ that is
yk = Hxk +vk
The random variables wk and kv represent the process and measurement noise respectively. They are assumed to be independent of each other, white, and with normal probability distributions p(w) ≈ N ,0( Q) (3) p(v) ≈ N ,0( R)
The more detailed working of Kalman filter can be found in [1].
Tracking Path of Moving Object
We provide you with two python scripts named tracker.py and kalman_filter.py. The scripts contain codes to track the object with its centroid. It makes use of Kalman filter algorithm. The codes are explained within the scripts with the help of comments.
tracker.py
# Import python libraries
import numpy as np
from kalman_filter import KalmanFilter
from common import dprint
from scipy.optimize import linear_sum_assignment
class Track(object):
"""Track class for every object to be tracked
Attributes:
None
"""
def __init__(self, prediction, trackIdCount):
"""Initialize variables used by Track class
Args:
prediction: predicted centroids of object to be tracked
trackIdCount: identification of each track object
Return:
None
"""
self.track_id = trackIdCount # identification of each track object
self.KF = KalmanFilter() # KF instance to track this object
self.prediction = np.asarray(prediction) # predicted centroids (x,y)
self.skipped_frames = 0 # number of frames skipped undetected
self.trace = [] # trace path
class Tracker(object):
"""Tracker class that updates track vectors of object tracked
Attributes:
None
"""
def __init__(self, dist_thresh, max_frames_to_skip, max_trace_length,
trackIdCount):
"""Initialize variable used by Tracker class
Args:
dist_thresh: distance threshold. When exceeds the threshold,
track will be deleted and new track is created
max_frames_to_skip: maximum allowed frames to be skipped for
the track object undetected
max_trace_lenght: trace path history length
trackIdCount: identification of each track object
Return:
None
"""
self.dist_thresh = dist_thresh
self.max_frames_to_skip = max_frames_to_skip
self.max_trace_length = max_trace_length
self.tracks = []
self.trackIdCount = trackIdCount
def Update(self, detections):
"""Update tracks vector using following steps:
- Create tracks if no tracks vector found
- Calculate cost using sum of square distance
between predicted vs detected centroids
- Using Hungarian Algorithm assign the correct
detected measurements to predicted tracks
https://en.wikipedia.org/wiki/Hungarian_algorithm
- Identify tracks with no assignment, if any
- If tracks are not detected for long time, remove them
- Now look for un_assigned detects
- Start new tracks
- Update KalmanFilter state, lastResults and tracks trace
Args:
detections: detected centroids of object to be tracked
Return:
None
"""
# Create tracks if no tracks vector found
if (len(self.tracks) == 0):
for i in range(len(detections)):
track = Track(detections[i], self.trackIdCount)
self.trackIdCount += 1
self.tracks.append(track)
# Calculate cost using sum of square distance between
# predicted vs detected centroids
N = len(self.tracks)
M = len(detections)
cost = np.zeros(shape=(N, M)) # Cost matrix
for i in range(len(self.tracks)):
for j in range(len(detections)):
try:
diff = self.tracks[i].prediction - detections[j]
distance = np.sqrt(diff[0][0]*diff[0][0] +
diff[1][0]*diff[1][0])
cost[i][j] = distance
except:
pass
# Let's average the squared ERROR
cost = (0.5) * cost
# Using Hungarian Algorithm assign the correct detected measurements
# to predicted tracks
assignment = []
for _ in range(N):
assignment.append(-1)
row_ind, col_ind = linear_sum_assignment(cost)
for i in range(len(row_ind)):
assignment[row_ind[i]] = col_ind[i]
# Identify tracks with no assignment, if any
un_assigned_tracks = []
for i in range(len(assignment)):
if (assignment[i] != -1):
# check for cost distance threshold.
# If cost is very high then un_assign (delete) the track
if (cost[i][assignment[i]] > self.dist_thresh):
assignment[i] = -1
un_assigned_tracks.append(i)
pass
else:
self.tracks[i].skipped_frames += 1
# If tracks are not detected for long time, remove them
del_tracks = []
for i in range(len(self.tracks)):
if (self.tracks[i].skipped_frames > self.max_frames_to_skip):
del_tracks.append(i)
if len(del_tracks) > 0: # only when skipped frame exceeds max
for id in del_tracks:
if id < len(self.tracks):
del self.tracks[id]
del assignment[id]
else:
dprint("ERROR: id is greater than length of tracks")
# Now look for un_assigned detects
un_assigned_detects = []
for i in range(len(detections)):
if i not in assignment:
un_assigned_detects.append(i)
# Start new tracks
if(len(un_assigned_detects) != 0):
for i in range(len(un_assigned_detects)):
track = Track(detections[un_assigned_detects[i]],
self.trackIdCount)
self.trackIdCount += 1
self.tracks.append(track)
# Update KalmanFilter state, lastResults and tracks trace
for i in range(len(assignment)):
self.tracks[i].KF.predict()
if(assignment[i] != -1):
self.tracks[i].skipped_frames = 0
self.tracks[i].prediction = self.tracks[i].KF.correct(
detections[assignment[i]], 1)
else:
self.tracks[i].prediction = self.tracks[i].KF.correct(
np.array([[0], [0]]), 0)
if(len(self.tracks[i].trace) > self.max_trace_length):
for j in range(len(self.tracks[i].trace) -
self.max_trace_length):
del self.tracks[i].trace[j]
self.tracks[i].trace.append(self.tracks[i].prediction)
self.tracks[i].KF.lastResult = self.tracks[i].predictionkalman_filter.py
import numpy as np
class KalmanFilter(object):
"""Kalman Filter class keeps track of the estimated state of
the system and the variance or uncertainty of the estimate.
Predict and Correct methods implement the functionality
Reference: https://en.wikipedia.org/wiki/Kalman_filter
Attributes: None
"""
def __init__(self):
"""Initialize variable used by Kalman Filter class
Args:
None
Return:
None
"""
self.dt = 0.005 # delta time
self.A = np.array([[1, 0], [0, 1]]) # matrix in observation equations
self.u = np.zeros((2, 1)) # previous state vector
# (x,y) tracking object center
self.b = np.array([[0], [255]]) # vector of observations
self.P = np.diag((3.0, 3.0)) # covariance matrix
self.F = np.array([[1.0, self.dt], [0.0, 1.0]]) # state transition mat
self.Q = np.eye(self.u.shape[0]) # process noise matrix
self.R = np.eye(self.b.shape[0]) # observation noise matrix
self.lastResult = np.array([[0], [255]])
def predict(self):
"""Predict state vector u and variance of uncertainty P (covariance).
where,
u: previous state vector
P: previous covariance matrix
F: state transition matrix
Q: process noise matrix
Equations:
u'_{k|k-1} = Fu'_{k-1|k-1}
P_{k|k-1} = FP_{k-1|k-1} F.T + Q
where,
F.T is F transpose
Args:
None
Return:
vector of predicted state estimate
"""
# Predicted state estimate
self.u = np.round(np.dot(self.F, self.u))
# Predicted estimate covariance
self.P = np.dot(self.F, np.dot(self.P, self.F.T)) + self.Q
self.lastResult = self.u # same last predicted result
return self.u
def correct(self, b, flag):
"""Correct or update state vector u and variance of uncertainty P (covariance).
where,
u: predicted state vector u
A: matrix in observation equations
b: vector of observations
P: predicted covariance matrix
Q: process noise matrix
R: observation noise matrix
Equations:
C = AP_{k|k-1} A.T + R
K_{k} = P_{k|k-1} A.T(C.Inv)
u'_{k|k} = u'_{k|k-1} + K_{k}(b_{k} - Au'_{k|k-1})
P_{k|k} = P_{k|k-1} - K_{k}(CK.T)
where,
A.T is A transpose
C.Inv is C inverse
Args:
b: vector of observations
flag: if "true" prediction result will be updated else detection
Return:
predicted state vector u
"""
if not flag: # update using prediction
self.b = self.lastResult
else: # update using detection
self.b = b
C = np.dot(self.A, np.dot(self.P, self.A.T)) + self.R
K = np.dot(self.P, np.dot(self.A.T, np.linalg.inv(C)))
self.u = np.round(self.u + np.dot(K, (self.b - np.dot(self.A,
self.u))))
self.P = self.P - np.dot(K, np.dot(C, K.T))
self.lastResult = self.u
return self.uNow, in the final part of the article, we will first draw tracking lines. Then, we draw bounding boxes for each object. The color of bounding box and tracking lines will be same and unique for each object.
We begin with importing Tracker class from the tracker.py script. We write the following code before the while loop i.e. before while cap.isOpened():-
from tracker import Tracker
traces = []
tracker = Tracker(200, 30, 20, 100)
# Variables initialization
skip_frame_count = 0
track_colors = [(255, 0, 0), (0, 255, 0), (0, 0, 255), (255, 255, 0),
(0, 255, 255), (255, 0, 255), (255, 127, 255),
(127, 0, 255), (127, 0, 127)]After adding this code, we proceed to write code within the while loop i.e. within while cap.isOpened() loop.
# If centroids are detected then track them
if len(centers) > 0:
# Track object using Kalman Filter
tracker.Update(centers)
# For identified object tracks draw tracking line
# Use various colors to indicate different track_id
colors = {}
for i in range(len(tracker.tracks)):
if len(tracker.tracks[i].trace) > 1:
for j in range(len(tracker.tracks[i].trace) - 1):
# Draw trace line
x1 = tracker.tracks[i].trace[j][0][0]
y1 = tracker.tracks[i].trace[j][1][0]
x2 = tracker.tracks[i].trace[j + 1][0][0]
y2 = tracker.tracks[i].trace[j + 1][1][0]
clr = tracker.tracks[i].track_id % 9
traces.append([x1,y1,x2,y2,clr])
for trace in traces:
frame = cv2.line(frame, (int(trace[0]), int(trace[1])), (int(trace[2]), int(trace[3])),
track_colors[trace[4]], 2)
if clr not in colors.keys():
colors[clr] = [x2, y2]
try:
c = []
for z in range(len(coords)):
minv = np.linalg.norm(np.array(centers[z]) - np.array(list(colors.values())[0]))
v = list(colors.keys())[0]
for val in range(1, len(colors.keys())):
dist = np.linalg.norm(np.array(centers[z]) - np.array(list(colors.values())[val]))
if dist < minv:
minv = dist
v = list(colors.keys())[val]
if v in c:
c.append(v + 1)
else:
c.append(v)
for z in range(len(coords)):
cv2.rectangle(frame, (coords[z][0], coords[z][1]), (coords[z][2], coords[z][3]),
track_colors[c[z]], 2)
except:
passThe above code block successfully draws tracking paths and bounding boxes on the frame. The frame can be shown using imshow() method.
Conclusion
In this article, we’ve examined how motion detection works in a video. We also worked with image processing techniques like blurring, diluting, finding differencesbetween frames. In addition, we saw how Kalman filter can be used to track the moving objects. At last, we learned about drawing bounding boxes and lines on the image with unique colors.
References
[1]. Mohamed LAARAIEDH. “Implementation of Kalman Filter with Python Language“. IETR Labs, University of Rennes 1. https://arxiv.org/ftp/arxiv/papers/1204/1204.0375.pdf